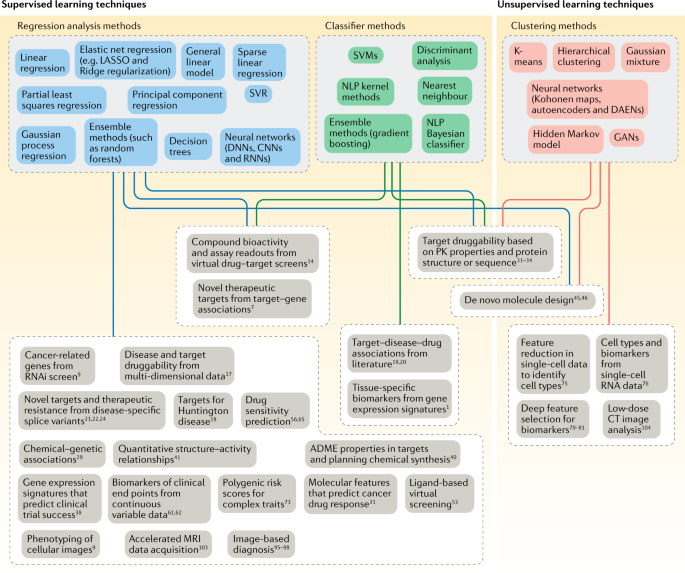
Drug Discovery
Youtube search... ...Google search News search...
- Case Studies
- COVID-19
- AI 'scientist' finds that toothpaste ingredient may help fight drug-resistant malaria
- FDA to Test New Trial Designs, Assess A.I. Models
- Allen Institute for Artificial Intelligence, or AI2’s SUPP.AI Discover Supplement-Drug Interactions
- Artificial Intelligence (AI) ... Generative AI ... Machine Learning (ML) ... Deep Learning ... Neural Network ... Reinforcement ... Learning Techniques
- Conversational AI ... ChatGPT | OpenAI ... Bing/Copilot | Microsoft ... Gemini | Google ... Claude | Anthropic ... Perplexity ... You ... phind ... Grok | xAI ... Groq ... Ernie | Baidu
- Generative AI will be designing new drugs all on its own in the near future | Trevor Jockims - CNBC ... “We now have this ability to represent the world of drugs — biology and chemistry — because we have AI supercomputers using AI and a GPT -like method, and with all of the digital biology data, we can represent the world of drugs in a computer for the very first time.”
Contents
Drug Discovery Development
Generative AI is transforming the drug discovery process.

Drug Discovery using Python
- ESOL: Estimating Aqueous Solubility Directly from Molecular Structure | John S. Delaney
- Maximizing computational tools for successful drug discovery | Chanin Nantasenamat and Virapong Prachayasittikul
- Machine Learning for Making Sense of Big Data in Drug Discovery Presentation | Chanin Nantasenamat
- Cheminformatics in Python: Predicting Solubility of Molecules | End-to-End Data Science Project Code | Chanin Nantasenamat - GitHub ...Libraries: PyCaret, scikit-learn, Pandas, and RDKit; a toolkit for cheminformatics. Environment: Colaboratory
- Using Computer Code to Decipher Genetic Code
- ROSALIND Platform
In this Jupyter notebook, we will dive into the world of Cheminformatics which lies at the interface of Informatics and Chemistry. We will be reproducing a research article (by John S. Delaney) by applying Linear Regression to predict the solubility of molecules (i.e. solubility of drugs is an important physicochemical property in Drug discovery, design and development). This idea for this notebook was inspired by the excellent blog post by Pat Walters where he reproduced the Linear Regression model with similar degree of performance as that of Delaney. This example is also briefly described in the book Deep Learning for the Life Sciences: Applying Deep Learning to Genomics, Microscopy, Drug Discovery, and More.Cheminformatics in Python: Predicting Solubility of Molecules | End-to-End Data Science Project | Chanin Nantasenamat
What can go wrong?
Network Biology
Network biology is a field of study that aims to understand the cell’s functional organization by systematically cataloging all molecules and their interactions within a living cell. It provides a new conceptual framework for understanding how these molecules and their interactions determine the function of this complex machinery. Network biology offers a quantifiable description of the networks that characterize various biological systems. There are numerous techniques used in network biology. Some of these techniques include high-throughput data-collection techniques that allow for simultaneous interrogation of the status of a cell’s components and determination of how and when these molecules interact with each other. Other techniques include bioinformatic techniques for genetic analysis using networks, based on random walks, information diffusion, and electrical resistance. These approaches have been applied successfully to identify disease genes, genetic modules, and drug targets. AI is being applied to network biology in various ways. One way is through the use of transfer learning, which leverages deep learning models pretrained on large-scale general datasets that can then be fine-tuned towards a vast array of downstream tasks with limited task-specific data. For example, a context-aware, attention-based deep learning model called Geneformer was developed and pretrained on a large-scale corpus of about 30 million single-cell transcriptomes to enable context-specific predictions in settings with limited data in network biology. This approach has been successful in accelerating the discovery of key network regulators and candidate therapeutic targets.

Gene-Network Biology
- Perspective ... Context ... In-Context Learning (ICL) ... Transfer Learning ... Out-of-Distribution (OOD) Generalization
- Causation vs. Correlation ... Autocorrelation ...Convolution vs. Cross-Correlation (Autocorrelation)
- Transfer learning enables predictions in network biology | C. Theodoris, L. Xiao, A. Chopra, M. Chaffin, Z. Sayed, M. Hill, H. Mantineo, E. Brydon, Z. Zeng, X. Liu & P. Ellinor ... Geneformer has been developed and pretrained using about 30 million single-cell gene-expression profiles to enable it to make predictions about gene-network biology in instances in which gene-expression data are limited.
Gene-Network Biology is a field of study that focuses on understanding the interactions between genes and how they determine the function of a cell. Geneformer can be tuned for many downstream applications to accelerate discovery of key gene-network regulators and candidate therapeutic targets. Network biology has served as a useful tool for the study of complex cellular systems by providing a glimpse into the functional organization of genes operating in normal and disease states. AI is being applied to Gene-Network Biology in various ways. One way is through the use of transfer learning, which leverages deep learning models pretrained on large-scale general datasets that can then be fine-tuned towards a vast array of downstream tasks with limited task-specific data. For example, a context-aware, attention-based deep learning model called Geneformer was developed and pretrained on a large-scale corpus of about 30 million single-cell transcriptomes to enable context-specific predictions in settings with limited data in network biology. This approach has been successful in accelerating the discovery of key network regulators and candidate therapeutic targets. Another way AI is being applied to Gene-Network Biology is through the use of machine learning platforms to work out patterns within large datasets that include many similar cells.

Gene Regulatory Networks (GRN)
- Gene regulatory network | Wikipedia
- Gene regulatory networks - Latest research and news | Nature
- ene Regulatory Network - an overview | ScienceDirect
- Gene Regulatory Networks: From DNA to development | Harvard
- Global gene network exploration based on explainable artificial intelligence approach | H. Park, K. Maruhashi, R. Yamaguchi, S. Imoto, S. Miyano - PLOS ONE
A Gene Regulatory Network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins. These interactions determine the function of the cell. GRNs also play a central role in morphogenesis, which is the creation of body structures. The regulators can be DNA, RNA, protein or any combination of two or more of these three that form a complex.
A simple example of a Gene Regulatory Network (GRN) could be one in which Gene A produces a protein that turns on Gene B, which itself produces a protein that turns on Gene C. This extra step allows for finer tuning in the levels of protein that Gene B and C produce. Another example of a GRN is one in which a yeast cell, finding itself in a sugar solution, will turn on genes to make enzymes that process the sugar to alcohol. This process is how the yeast cell makes its living, gaining energy to multiply. Artificial Intelligence (AI) has been applied to Gene Regulatory Networks (GRNs) in various ways. One approach is to use an interpretable AI method based on tensor decomposition to overcome limitations of existing gene network analysis and reveal global and novel mechanisms of gene regulatory systems from massive multiple networks. Another approach is to use deep learning models to filter noise in single-cell transcriptome data by modeling the complicated interaction patterns among genes. These deep learning-based methods are able to model gene interactions to reveal a clearer landscape of cell heterogeneity.
DeepSEM, a deep generative model, that can jointly infer Gene Regulatory Networks (GRNs) and biologically meaningful representation of single-cell RNA sequencing (scRNA-seq) data . They developed a neural network version of the structural equation model (SEM) to explicitly model the regulatory relationships among genes . Benchmark results show that DeepSEM achieves comparable or better performance on a variety of single-cell computational tasks compared with state-of-the-art methods. [Modeling gene regulatory networks using neural network architectures | H. Shu, J. Zhou, Q. Lian, H. Li, D. Zhao, J. Zeng & J. Ma - Nature] ... DeepSEM | GitHub

Protein Interaction Networks
Protein Interaction Networks are networks of protein complexes formed by biochemical events and/or electrostatic forces that serve a distinct biological function as a complex. AI has been applied to Protein Interaction Networks in various ways. For example, researchers at UT Southwestern and University of Washington led an international team that used AI and evolutionary analysis to produce 3D models of eukaryotic protein interactions1. To find proteins that were likely to interact, the scientists first searched the genomes of related fungi for genes that acquired mutations in a linked fashion. They then used two AI technologies to determine whether these proteins could be fit together in 3D structures
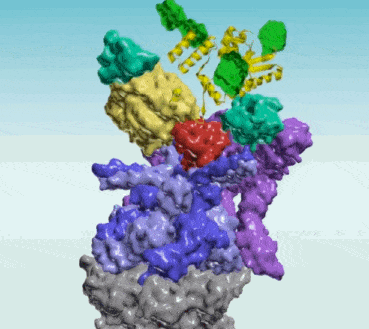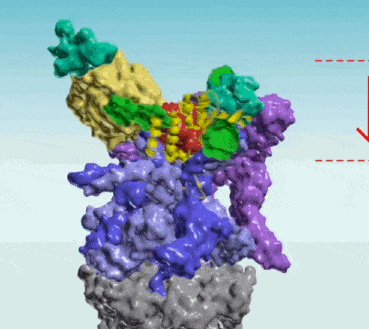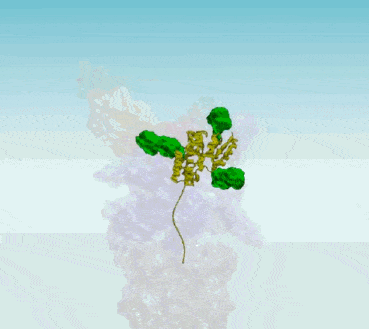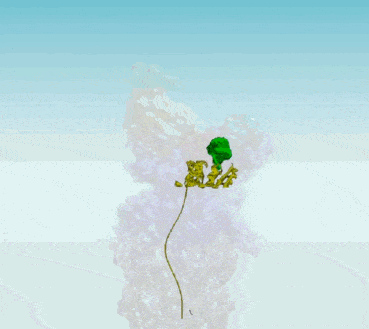
Protein Protein Interaction Networks (PPI)
- Protein-Protein Interaction Networks | Marco Pellegrini - ScienceDirect ... PPI network alignment can be treated as a supplement to the computation of sequence similarity, providing topological information that may be needed to identify orthologous proteins.
It is hard to think of a biological process in which protein-protein interactions (PPIs) do not play an essential role. Thus, in collective efforts over the last two decades comprehensive sets of human PPIs have been curated from the scientific literature or identified in systematic, proteome-wide mapping efforts. These resources build large PPI networks with an amazing potential to advance our understanding of individual gene function towards a systems understanding of cellular organization. When using protein-protein interaction (PPI) data for system-wide analyses, it is important to consider technical biases that may affect the results. Research will provide important insights into these biases and explain the theory and practical considerations when performing statistical tests on PPI networks. One key consideration is determining whether selected proteins, such as those that share a disease association, tend to interact with each other. This can be done through statistical tests on PPI networks. Additionally, the prediction of gene function can be done using guilt-by-association principles. By understanding these technical biases and considerations, researchers can more accurately analyze PPI data and draw meaningful conclusions from their results.
There are many online resources for Protein-Protein Interaction Networks (PPI). Some of these include the Database of Interacting Protein (DIP), the Biomolecular Interaction Network Database (BIND), the Münich Information Center for Protein Sequence (MIPS) protein interaction database, the Molecular Interaction database (MINT), the protein Interaction database (IntAct), the Biological General Repository for Interaction Datasets (BioGRID), and the Human Protein Reference Database (HPRD).

Dual Use
- Dual use of artificial-intelligence-inspired drug discovery | F. Urbina, F. Lentzos, C. Invernizzi & S. Ekins - Nature Machine Intelligence
- Drug-discovery AI can be inverted to create chemical weapons, scientists find | Dan McCarthy - Tech Brew ... It took researchers under six hours to coax a drug-discovery AI into producing 40,000 toxic molecules.
The dual use of artificial intelligence in drug discovery refers to the potential for AI-powered drug discovery tools to be misused for the de novo design of biochemical weapons.
The paper "Dual use of artificial-intelligence-inspired drug discovery" discusses the potential for harm in using cutting-edge technology in drug discovery. The researchers use a machine learning "molecule generator" called "MegaSyn" to generate compounds that can be used as a drug. However, they also demonstrated that the system's goals could be reversed, and it could be used to generate biowarfare agents. The paper reflects a group of drug developers reckoning with the potential for harm inherent in their cutting-edge technology. The researchers demonstrated how even tools designed for good, such as AI-powered drug discovery, could be tweaked to produce dire results. MegaSyn can generate thousands of novel chemical weapons in a matter of hours.
The dual use of artificial intelligence in drug discovery has been a topic of concern among scientists. Here are some examples of how AI technologies can be misused for the de novo design of biochemical weapons:
- Thought experiment and computational proof: An international security conference explored the dual-use potential of AI in drug discovery, which evolved from a thought experiment into a computational proof. This demonstrated the alarming results of using AI approaches for the design of nerve agents, including VX.
- Generative AI approach for nerve agents: Researchers used a generative AI approach previously developed for drug discovery applications and found that it could easily design a range of nerve agents, highlighting the potential for dual use.
- Time to consider dual-use risk: The results of the computational experiment have led to a call for considering the dual-use risk of the development of toxic agents in silico, in addition to physical biological agents.
- Teachable moment for dual-use: The thought experiment has become a "teachable moment for dual-use," providing a test case for considering the risks of research involving converging technologies.
- Dual-use risk training: The study can be used to provide dual-use risk training for those applying AI in drug discovery, specifically in the context of nerve agents and chemical weapons.
- Alarming potential for dual use: The collaboration between researchers found that the potential for dual use in this case is alarmingly high, highlighting the need for further exploration and mitigation of these risks.