Protein Folding & Discovery
Youtube search... ...Google search
- Symbiotic Intelligence ... Bio-inspired Computing ... Neuroscience ... Connecting Brains ... Nanobots ... Molecular ... Neuromorphic ... Evolutionary/Genetic
- Case Studies
- COVID-19
- Service Capabilities
- Architectures for AI ... Enterprise Architecture (EA) ... Enterprise Portfolio Management (EPM) ... Architecture and Interior Design
- (Deep) Residual Network (DRN) - ResNet
- RoseTTAFold: Accurate protein structure prediction accessible to all | Institute for Protein Design - University of Washington
- AI Reveals Previously Unknown Biology – We Might Not Know Half of What’s in Our Cells | University of California-San Diego
- Artificial intelligence powers protein-folding predictions | Michael Eisenstein - Nature
- Artificial intelligence has worked out the structures of 200 million proteins (that’s practically all of them) | Stephanie Pappas - Livescience
Proteins are made up of hundreds or thousands of amino acids, and these amino acid sequences specify the protein's structure and function. But understanding just how to build these sequences to create novel proteins has been challenging. Past work has resulted in methods that can specify structure, but function has been more elusive. Machine learning reveals recipe for building artificial proteins | Emily Ayshford - University of Chicago - PhysOrg
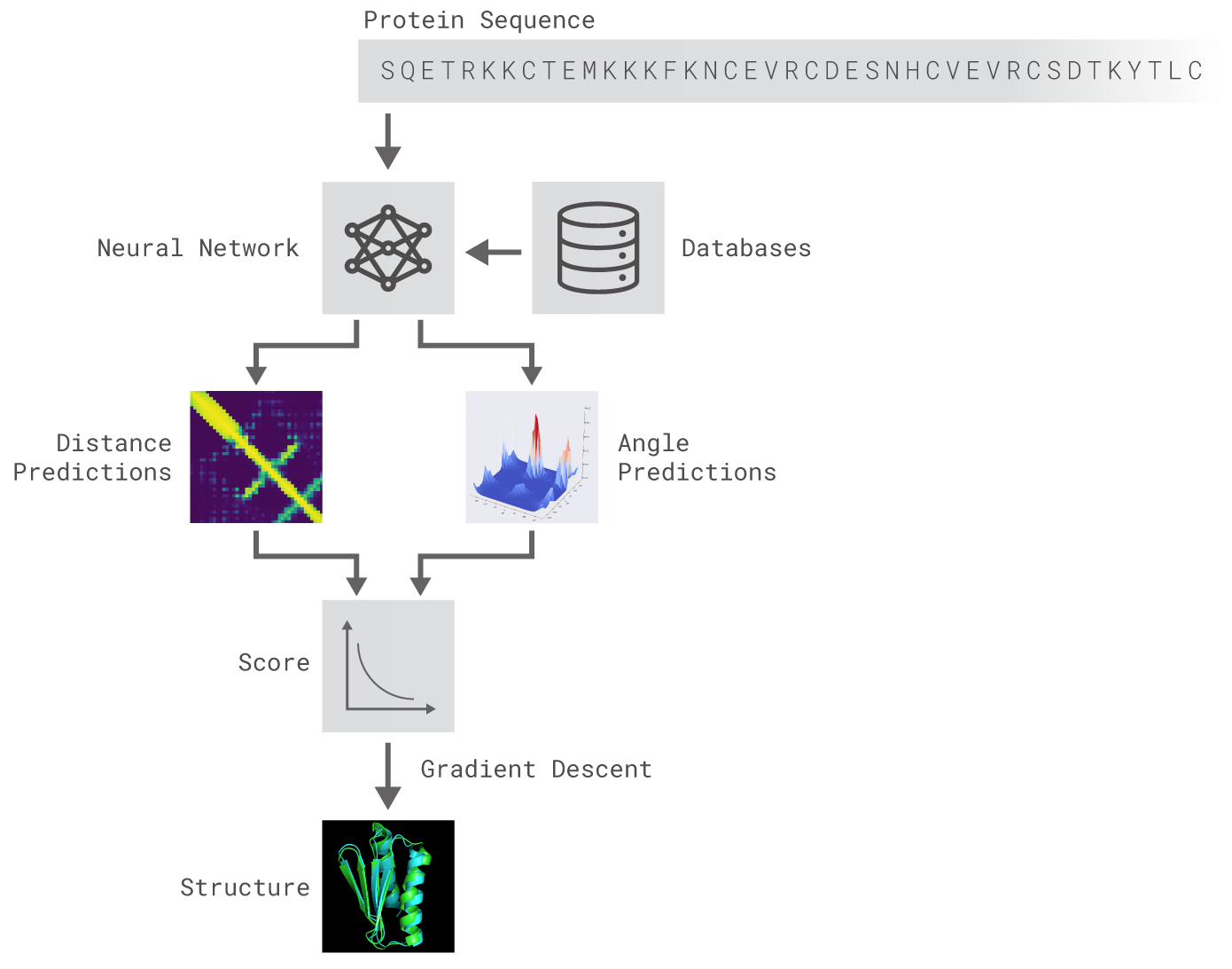
What is the protein folding problem? Proteins are large, complex molecules essential to all of life. Nearly every function that our body performs—contracting muscles, sensing light, or turning food into energy—relies on proteins, and how they move and change. What any given protein can do depends on its unique 3D structure. For example, antibody proteins utilised by our immune systems are ‘Y-shaped’, and form unique hooks. By latching on to viruses and bacteria, these antibody proteins are able to detect and tag disease-causing microorganisms for elimination. Collagen proteins are shaped like cords, which transmit tension between cartilage, ligaments, bones, and skin. Other types of proteins include Cas9, which, using CRISPR sequences as a guide, act like scissors to cut and paste sections of DNA; antifreeze proteins, whose 3D structure allows them to bind to ice crystals and prevent organisms from freezing; and ribosomes, which act like a programmed assembly line, helping to build proteins themselves. The recipes for those proteins—called genes—are encoded in our DNA. An error in the genetic recipe may result in a malformed protein, which could result in disease or death for an organism. Many diseases, therefore, are fundamentally linked to proteins. But just because you know the genetic recipe for a protein doesn’t mean you automatically know its shape. Proteins are comprised of chains of amino acids (also referred to as amino acid residues). But DNA only contains information about the sequence of amino acids–not how they fold into shape. The bigger the protein, the more difficult it is to model, because there are more interactions between amino acids to take into account. AlphaFold: Using AI for scientific discovery | A. Senior, J. Jumper, D. Hassabis, and P. Kohli - DeepMind
Google DeepMind AlphaFold
Youtube search... ...Google search
- Google DeepMind AlphaFold
- Google DeepMind AlphaGo Zero ... * Google DeepMind AlphaStar
- 13th Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction
- Protein Data Bank | Wikipedia
- AlphaFold: a solution to a 50-year-old grand challenge in biology | DeepMind
- AI has cracked a problem that stumped biologists for 50 years. It’s a huge deal. | Sigal Samuel - Vox
- DeepMind and European Molecular Biology Laboratory (EMBL) release the most complete database of predicted 3D structures of human proteins | SciTechDaily Partners use AlphaFold, the AI system recognized last year as a solution to the protein structure prediction problem, to release more than 350,000 protein structure predictions including the entire human proteome to the scientific community.
- Highly accurate protein structure prediction with AlphaFold | J. Jumper, R. Evans, D. Hassabis - Nature
- FastFold: Reducing AlphaFold Training Time from 11 Days to 67 Hours | S. Cheng, R. Wu, Z. Yu, B. Li, X. Zhang, J. Peng, Y. You
- Google DeepMind Introduces AlphaFold 3: A Revolutionary AI Model that can Predict the Structure and Interactions of All Life’s Molecules with Unprecedented Accuracy | Asif Razzaq - MarketTechPost
DeepMind has brought together experts from the fields of structural biology, physics, and machine learning to apply cutting-edge techniques to predict the 3D structure of a protein based solely on its genetic sequence. AlphaFold: Using AI for scientific discovery | DeepMind
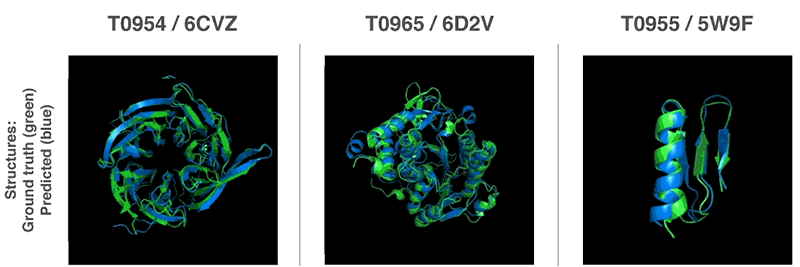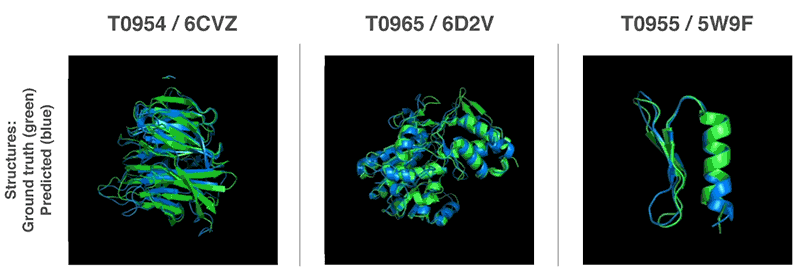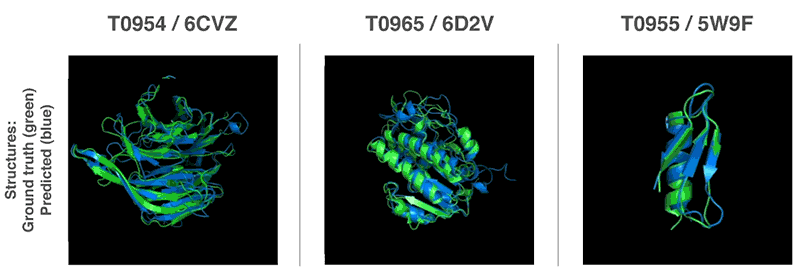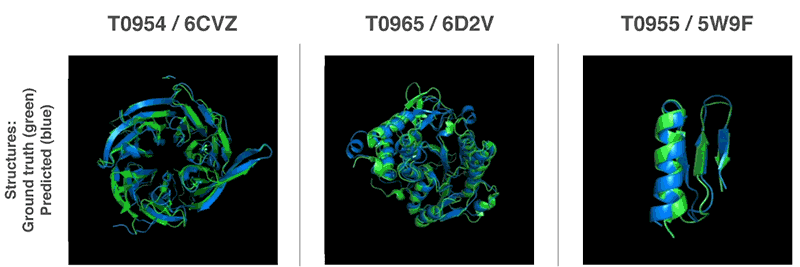
AlphaFold Demo
Google DeepMind and Isomorphic Labs have announced that scientists will have free access to most features of their newly launched research tool, the AlphaFold Server.
To use the AlphaFold server demo, you need to start by accessing the AlphaFold server website: https://AlphaFoldServer.com
Once there, you may need to create an account or log in if you haven't already.
Next, prepare your input data by creating a FASTA file that contains the amino acid sequence of the protein you wish to analyze. The FASTA format is a simple text-based format for representing nucleotide or peptide sequences, where each sequence is preceded by a description line starting with a ">" character. Make sure your FASTA file is correctly formatted and accurately represents the protein sequence.
Once your FASTA file is ready, go to the web interface of the AlphaFold server. Here, you will find an option to upload your FASTA file, usually through a clear "Upload" button. After uploading the file, you may need to provide additional details about the protein or adjust specific parameters for the prediction. This step ensures that the AI model has all the necessary information to process your job accurately.
After submitting the FASTA file and any necessary information, you can start the prediction job by clicking the button to begin the process, typically labeled "Submit," "Run," or something similar. The processing time can vary depending on the complexity of the protein and the current server load. You might see a progress bar or receive a notification once the job is complete.
When the prediction is finished, the server will provide an overview of the protein structure as a 3D rendering. This rendering is usually interactive, allowing you to manipulate and explore the 3D model directly within your web browser. Additionally, there should be options to download the predicted structure in various formats, such as PDB, for further analysis or use in other bioinformatics tools.
Finally, you can utilize the predicted structure for your research purposes. This might involve conducting further computational analysis, planning laboratory experiments, or integrating the structure into scholarly work. The streamlined process offered by the AlphaFold 3 server demo makes it significantly easier for researchers to obtain high-quality protein structure predictions and apply these insights to their scientific endeavors.
In summary, using the AlphaFold 3 server involves accessing the web interface, preparing and uploading a FASTA file, submitting the job, waiting for the prediction to complete, and then reviewing and utilizing the results. This user-friendly approach enables researchers to conduct sophisticated protein structure analysis with minimal effort, supporting a wide range of biological and biomedical research projects.
FASTA & FASTQ
FASTA and FASTQ are two widely used file formats in bioinformatics for storing nucleotide or peptide sequences. Here’s a detailed explanation of each:
FASTA is used to represent nucleotide sequences or protein sequences. The structure of a FASTA file includes a header line that begins with a '>' character followed by a description (e.g., sequence name or identifier), and sequence lines that contain the actual nucleotide or protein sequence in a single-letter code, which can span multiple lines. For example:
``` >sequence_1 ATGCGTAACGTAGCTAGCTAGCTAGCTA GCTAGCTAGCATCGATCGATCGATCGA ```
FASTQ is used to store both nucleotide sequences and their corresponding quality scores, which indicate the confidence of each base call, commonly produced by high-throughput sequencing technologies. The structure of a FASTQ file includes a header line that begins with an '@' character followed by a description (e.g., sequence name or identifier), a sequence line that contains the actual nucleotide sequence, a plus line with a '+' character (sometimes followed by the same identifier as in the header line, optionally), and a quality line with encoded quality scores corresponding to each nucleotide in the sequence line. The length of the quality line matches the length of the sequence line. For example:
``` @sequence_1 ATGCGTAACGTAGCTAGCTAGCTAGCTA + IIIIIIIIIIIIIIIIIIIIIIIIIIII ```
The key differences between these formats are that FASTA stores only the sequence information, while FASTQ stores both sequence information and quality scores. FASTA is typically used for sequence alignment, database searches, and various other bioinformatics analyses due to its simplicity and lower data intensity compared to FASTQ. On the other hand, FASTQ is essential in next-generation sequencing (NGS) workflows where quality scores are critical for downstream processing like read mapping, variant calling, and quality filtering, making it more complex due to the inclusion of quality scores.
Quality scores in FASTQ files are typically encoded using ASCII characters, with different versions of FASTQ (e.g., Sanger, Illumina 1.8+) using different offsets for encoding these scores. For instance, the Sanger format uses ASCII 33-73 to represent Phred quality scores ranging from 0 to 40. Understanding these formats is crucial for tasks such as sequence assembly, annotation, and various genomic analyses in bioinformatics.