Protein Folding & Discovery
Youtube search... ...Google search
- Case Studies
- COVID-19
- Service Capabilities
- Evolutionary Computation / Genetic Algorithms
- Architectures
- (Deep) Residual Network (DRN) - ResNet
Proteins are made up of hundreds or thousands of amino acids, and these amino acid sequences specify the protein's structure and function. But understanding just how to build these sequences to create novel proteins has been challenging. Past work has resulted in methods that can specify structure, but function has been more elusive. Machine learning reveals recipe for building artificial proteins | Emily Ayshford - University of Chicago - PhysOrg
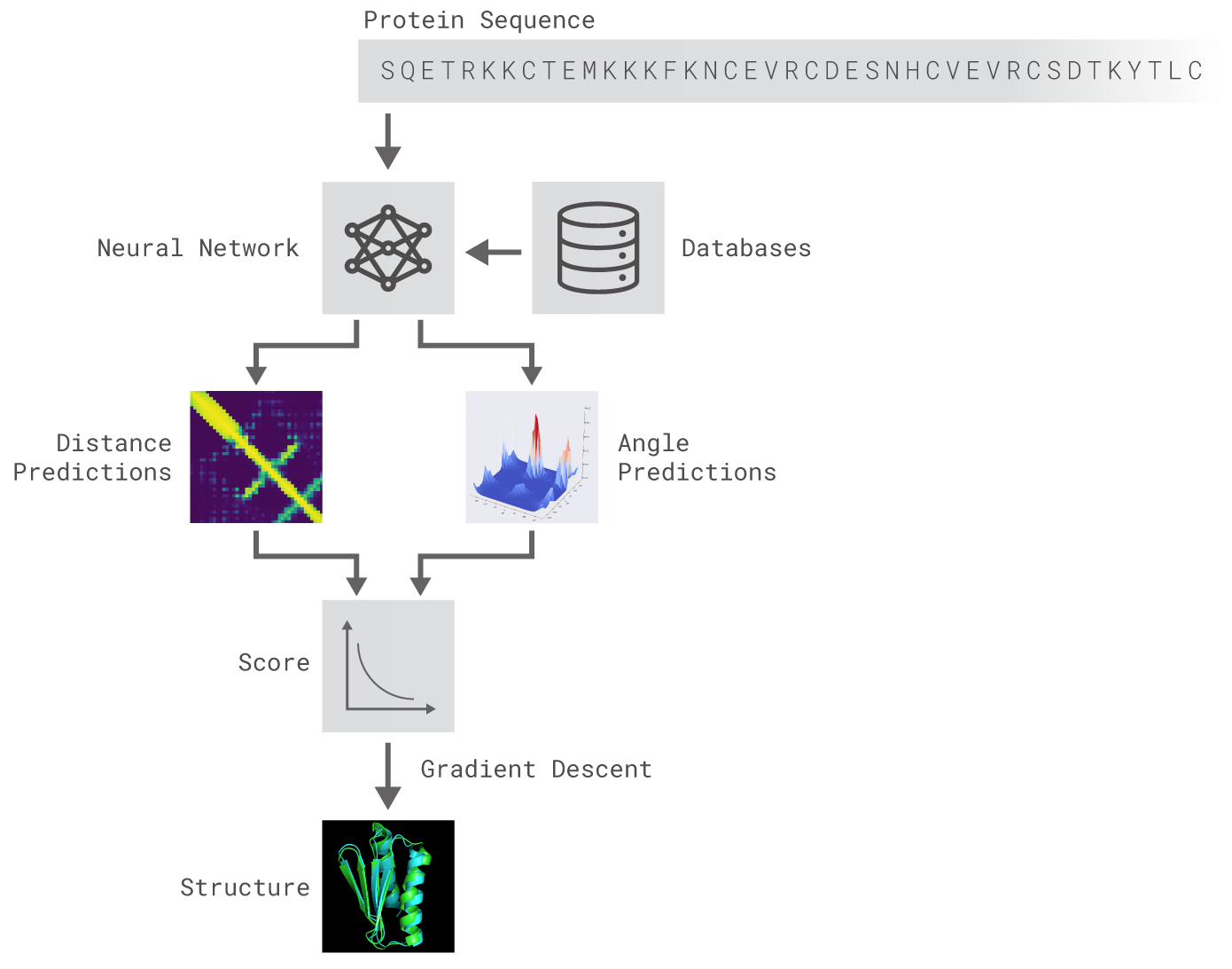
What is the protein folding problem? Proteins are large, complex molecules essential to all of life. Nearly every function that our body performs—contracting muscles, sensing light, or turning food into energy—relies on proteins, and how they move and change. What any given protein can do depends on its unique 3D structure. For example, antibody proteins utilised by our immune systems are ‘Y-shaped’, and form unique hooks. By latching on to viruses and bacteria, these antibody proteins are able to detect and tag disease-causing microorganisms for elimination. Collagen proteins are shaped like cords, which transmit tension between cartilage, ligaments, bones, and skin. Other types of proteins include Cas9, which, using CRISPR sequences as a guide, act like scissors to cut and paste sections of DNA; antifreeze proteins, whose 3D structure allows them to bind to ice crystals and prevent organisms from freezing; and ribosomes, which act like a programmed assembly line, helping to build proteins themselves. The recipes for those proteins—called genes—are encoded in our DNA. An error in the genetic recipe may result in a malformed protein, which could result in disease or death for an organism. Many diseases, therefore, are fundamentally linked to proteins. But just because you know the genetic recipe for a protein doesn’t mean you automatically know its shape. Proteins are comprised of chains of amino acids (also referred to as amino acid residues). But DNA only contains information about the sequence of amino acids–not how they fold into shape. The bigger the protein, the more difficult it is to model, because there are more interactions between amino acids to take into account. AlphaFold: Using AI for scientific discovery | A. Senior, J. Jumper, D. Hassabis, and P. Kohli - DeepMind
Google DeepMind AlphaFold
Youtube search... ...Google search
- Google DeepMind AlphaGo Zero
- Google DeepMind AlphaStar
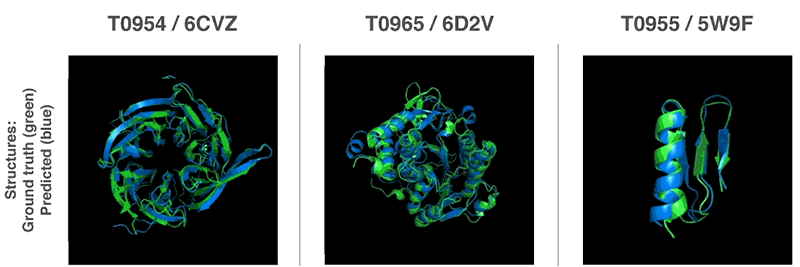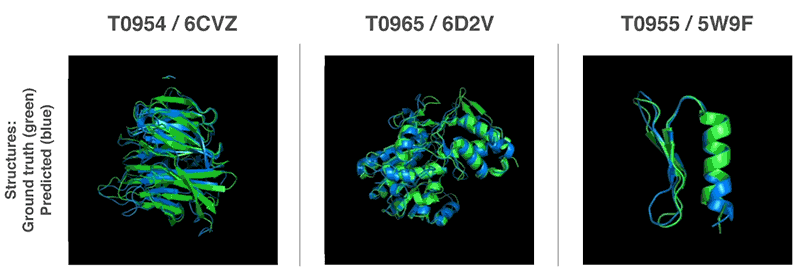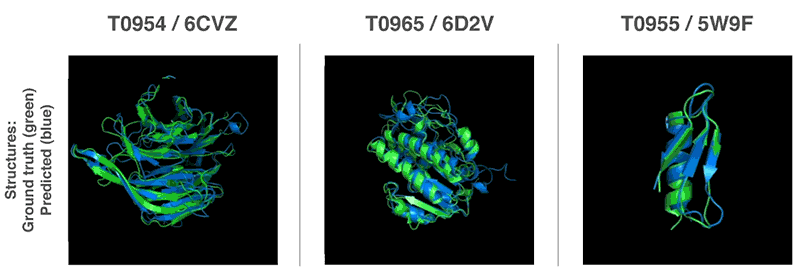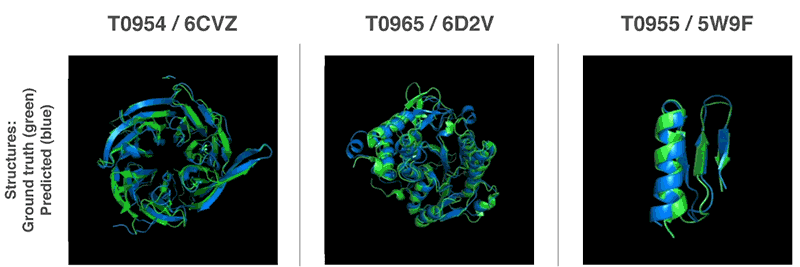
- 13th Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction
- Protein Data Bank | Wikipedia
- AI has cracked a problem that stumped biologists for 50 years. It’s a huge deal. | Sigal Samuel - Vox
- [DeepMind and European Molecular Biology Laboratory (EMBL) release the most complete database of predicted 3D structures of human proteins | SciTechDaily Partners use AlphaFold, the AI system recognized last year as a solution to the protein structure prediction problem, to release more than 350,000 protein structure predictions including the entire human proteome to the scientific community.
DeepMind has brought together experts from the fields of structural biology, physics, and machine learning to apply cutting-edge techniques to predict the 3D structure of a protein based solely on its genetic sequence. AlphaFold: Using AI for scientific discovery | DeepMind